
We published our scallop genome paper on Nature Ecology & Evolution
On April 3, 2017, Nature Ecology & Evolution published the latest paper "Scallop genome provides insights into evolution of bilaterian karyotype and development" online in Article form by an international research team led by Professor Zhenmin Bao from the Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China. Our group, in cooperation with researchers from Novogene (Beijing), Sars International Centre (Norway), Rutgers University (USA) and Institute of Oceanology, Chinese Academy of Sciences, has completed the first fine-mapping of the scallop genome in the world and made important progress in the field of animal macroevolution, making several new discoveries and insights into the evolution of chromosome karyotype of Urbilateria, the generation of body plan diversity, and the origin and regulatory mechanisms of the eye, which provide key clues for understanding the origin and evolution mechanism of early animals.
On the earth today, 99% of the species in the animal kingdom belongs to bilaterian, which emerged in large numbers during the Cambrian explosion and evolved to form the basic pattern of the major taxa of the animal kingdom on our planet today. Evolutionists have speculated that these bilaterians may have originated from an ancient common ancestor (Urbilateria), but due to the paucity of early fossil records, the origin and evolutionary mechanisms of bilaterians have long been controversial. As the initiator of the International Scallop Genome Project, Prof. Zhenmin Bao's group has led the first high-quality genome-wide mapping of scallops. We assembled the ancient lophotrochozoan genome at the chromosome level for the first time using our previously developed 2b-RAD technology (Nature Methods, 2012). The deep analysis of the scallop genome reveals that scallops exhibit many original ancestral genomic features, such as nearly perfect preservation of the ancestral karyotype, the largest number of ancient gene families, and the most complete Hox, ParaHox, and NK gene clusters, which are the oldest bilaterally symmetrical animal genomes ever discovered. Shellfish appeared in the Early Cambrian 500 million years ago, and they provide a valuable genomic model for studying the evolution of bilaterian. Inferring genomic features of Urbilateria by reconstruction may provide key clues for understanding the mechanisms of early animal origins and evolution.
The study further reveals that the expression pattern of the Hox gene cluster controlling body plan development in scallops has a subcluster-level temporal co-linearity (STC) expression pattern, which is different from the current consensus that the complete Hox gene cluster follows whole-cluster temporal co-linearity (WTC) expression pattern. In-depth studies reveal that the STC expression pattern is common in lower animals and is a unique and ancient body pattern determined mechanism, which provides a new perspective for understanding the origin and evolutionary mechanism of the diversification of body patterns in Cambrian animals.
Scallops have tens to hundreds of mirror eyes, and the study also revealed that scallop eyes possess an unusual multiple phototransduction pathway systems, and their eye development is dominated by the pax2/5/8 gene rather than the pax6 gene, challenging the currently prevailing international hypothesis of a monogenic origin of the eye (controlled by pax6) and providing key evidence for the new hypothesis that the lateral eyes of the animal body evolved independently from the origin of the cephalic eye. The above important findings elevate shellfish to a new status in the field of animal evolution research, expanding and deepening the current academic understanding of the early origin and evolutionary mechanisms of bilaterians, and exploring the origin and evolution of animals is a major scientific question in the field of life.
Prof. Zhenmin Bao and Prof. Shi Wang of the Key Laboratory of Marine Biogenetics and Breeding of the Ministry of Education were the corresponding authors and the first author of the paper, respectively. The research work was funded by the National Natural Science Foundation of China, the National High Technology Research and Development Program of China (863 program), and the AoShan Talents Program of Qingdao National Laboratory for Marine Science and Technology. Dalian Zhangzidao Group also provided strong support for the successful completion of the project with funds and materials.
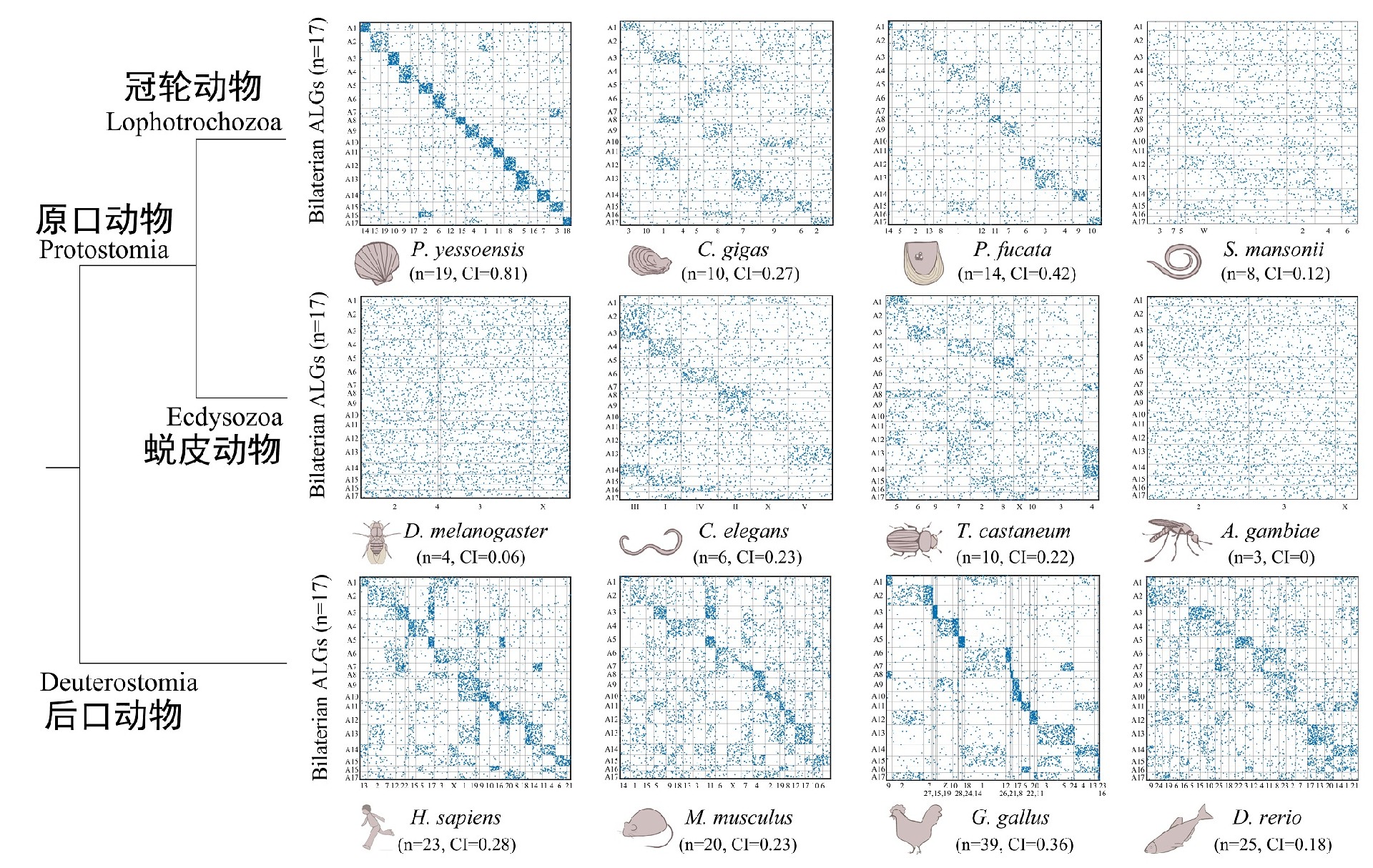
Figure 1. The scallop (P. yessoensis) almost perfectly retains the chromosome karyotype of Urbilateria
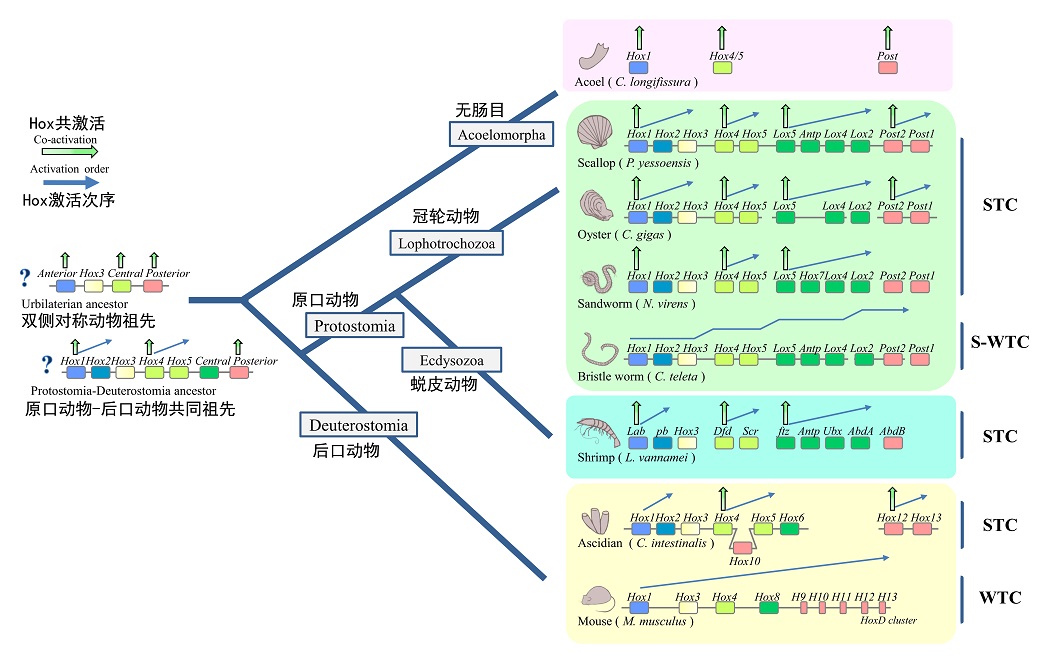
Figure 2. Novel patterns of "subcluster-level temporal co-linearity (STC)" regulation of Hox gene clusters and its possible origin and evolutionary pathway
Paper links: http://www.nature.com/articles/s41559-017-0120

